 How to determine HRD status?
How to determine HRD status?
When the homologous recombination (HR) pathway is disrupted by gene mutations, promoter methylation, or undetermined causes, the HR pathway stops working, leading to Homologous Recombination Deficiency (HRD).
Tumors with HRD cannot repair themselves effectively after sustaining damage, contributing to genomic instability. There are limitations to determining HRD status when evaluating each 'cause'/ cause individually. Evaluating genomic instability allows for the assessment of HRD regardless of the specific cause.
HRD status can be measured by “cause” through mutations in the HRR pathway (e.g., BRCA1 and BRCA2) and by the “effect” of the presence of genomic scars at a given threshold or functional assay. HRD testing can identify 30% more PARPi-effective population than BRCA testing. (BRCA~20% vs. HRD~50%)
AmoyDx® HRD Complete Panel offers a comprehensive homologous recombination deficiency (HRD) test designed to simultaneously detect genetic abberations across 20 HRR genes and HRD status.

Comprehensively Designed GSS Algorithm
The AmoyDx proprietary GSS algorithm is a machine learning-based model which assesses genomic instability by analyzing different types of copy number events across the genome.

Longer PFS with PARPi Treatment for GSS-positive Group
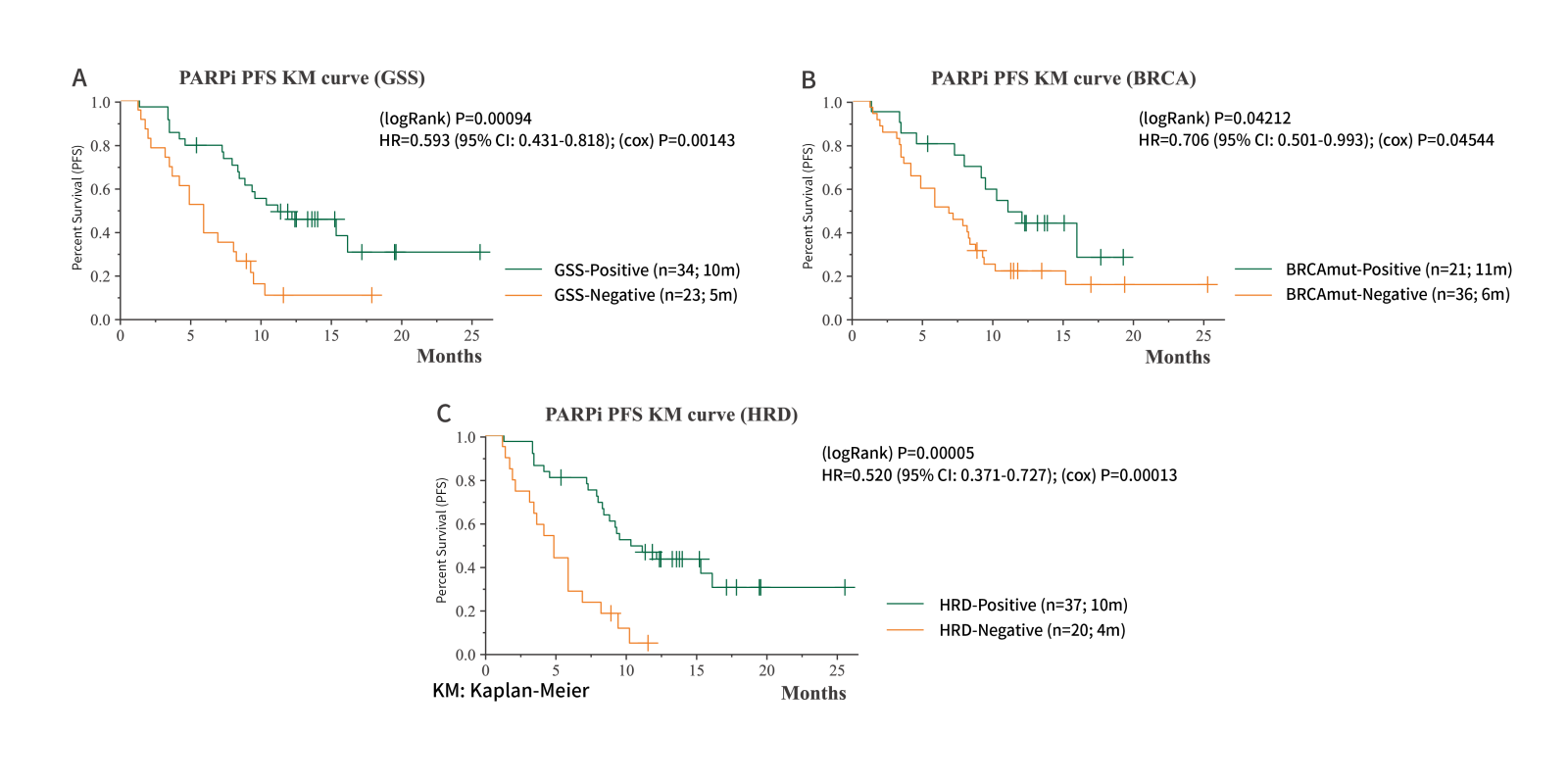 The study highlights the promising value of GSS in identifying patients
The study highlights the promising value of GSS in identifying patients who may respond favorably to PARPi treatment.
White Paper
1. Comparative Analysis of HRD Complete Data on MGI DNBSEQ-G50 and NextSeq 550
The study confirms that the AmoyDx® HRD Complete Panel performs exceptionally well on both MGI G50 and NextSeq 550 platforms. The MGI G50 demonstrated superior sequencing quality and data QC performance while maintaining comparable accuracy in SNVs/InDels and HDs detection as well as HRD (GSS) assessment. These findings validate MGI G50 as a robust and reliable platform for HRR gene mutation screening and HRD (GSS) status analysis.Mutations( Hover over each mutational type to highlight genes covered )
All

ATM
ATM
HD
SNV/INDELS
BARD1
BARD1
HD
SNV/INDELS
BRCA1
BRCA1
HD
SNV/INDELS
BRCA2
BRCA2
HD
SNV/INDELS
BRIP1
BRIP1
HD
SNV/INDELS
CDH1
CDH1
HD
SNV/INDELS
CDK12
CDK12
HD
SNV/INDELS
CHEK1
CHEK1
HD
SNV/INDELS
CHEK2
CHEK2
HD
SNV/INDELS
FANCA
FANCA
HD
SNV/INDELS
FANCL
FANCL
HD
SNV/INDELS
HDAC2
HDAC2
HD
SNV/INDELS
PALB2
PALB2
HD
SNV/INDELS
PPP2R2A
PPP2R2A
HD
SNV/INDELS
PTEN
PTEN
HD
SNV/INDELS
RAD51B
RAD51B
HD
SNV/INDELS
RAD51C
RAD51C
HD
SNV/INDELS
RAD51D
RAD51D
HD
SNV/INDELS
RAD54L
RAD54L
HD
SNV/INDELS
TP53
TP53
HD
SNV/INDELS

Specifications
Alterations detected
20 HRR genes (SNV/InDel/HD) and Genomic Scar Score (GSS)
Sample type
FFPE tissue
DNA input
Optimal 100 ng (minimum 50 ng)
Data output per sample
4 Gb
Sequencing type
PE150
Sequencer
Illumina NextSeq 500/550, NovaSeq 6000
TAT for library preparation
5 hours (hands-on time <1 hour)
TAT from sample to report
3 days
Publications


1. Feng, Cong et al. Relationship between homologous recombination deficiency and clinical features of breast cancer based on genomic scar score. The Breast, Volume 69, 392 - 400.
2. Yuan W, Ni J, Wen H, Shi W, Chen X, Huang H, et al. Genomic Scar Score: A robust model predicting homologous recombination deficiency based on genomic instability. BJOG. 2022; 129(Suppl. 2): 14–22.
Inquiry





